Chapter 6 RNA-seq FDA benchmark paper
Title Impact of RNA-seq data analysis algorithms on gene expression estimation and downstream prediction
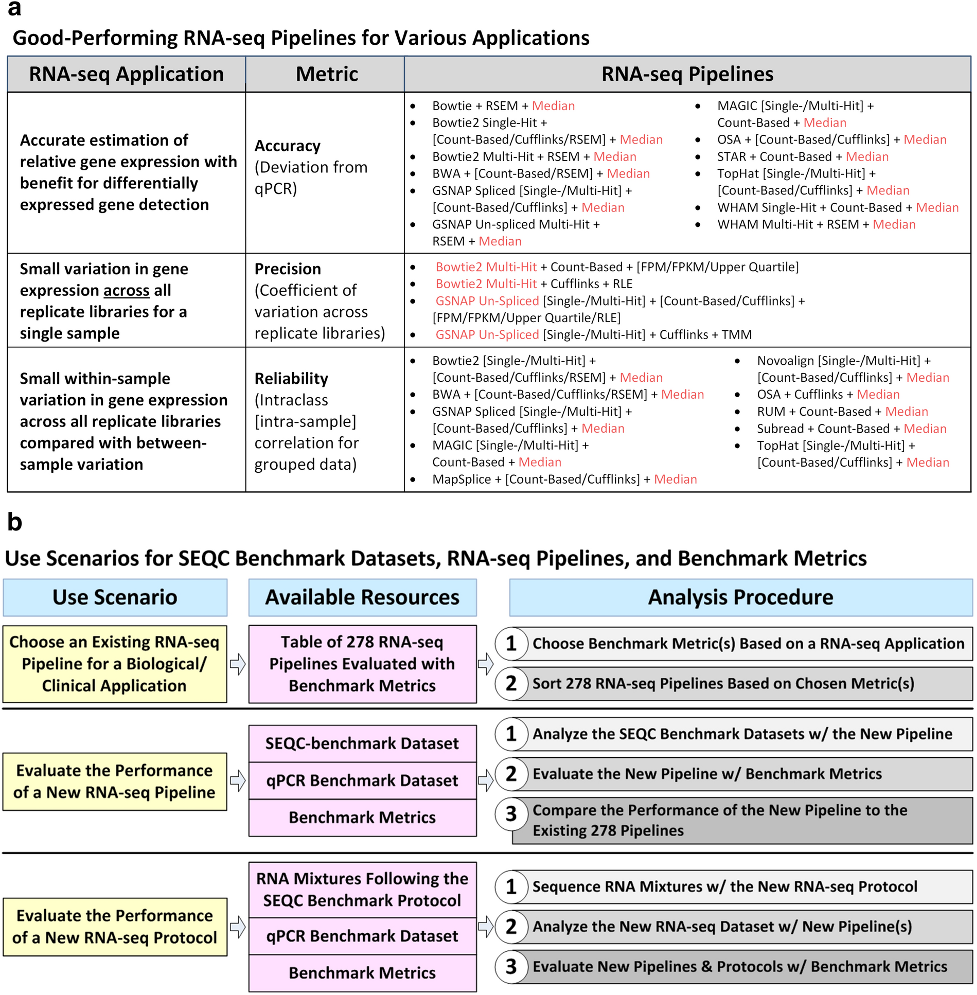
Results
Accuracy - as the deviation of RNA-seq pipeline-derived log ratios of gene expression from the corresponding qPCR-based log ratios. A smaller deviation represents higher accuracy. Median normalization exhibited the lowest deviation, or the highest accuracy, compared with all other normalization methods.
Precision - as the coefficient of variation (CoV) of gene expression across replicate libraries. Smaller CoV represents higher precision. pipelines with any of Bowtie2 multi-hit, GSNAP un-spliced, or Subread mapping and either count-based or Cufflinks quantification, except for the [Bowtie2 multi-hit + count-based + med.] pipeline, were the best choice for quantifying genes with high precision, or low CoV.
Reliability - as the intra-class (i.e., intra-sample in the context of the SEQC-benchmark dataset) correlation (ICC) of gene expression. Larger ICC represents higher reliability. median normalization along with most mapping and quantification algorithms, except for the [Bowtie2 multi-hit + count-based] and [Novoalign + RSEM] pipelines, was the best choice for quantifying genes with high reliability, or high ICC.
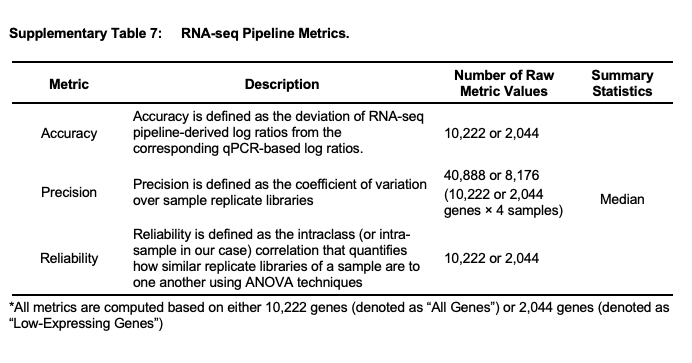 * The resulting heatmaps in accrodance to pipeline metrics strategy
* The resulting heatmaps in accrodance to pipeline metrics strategy 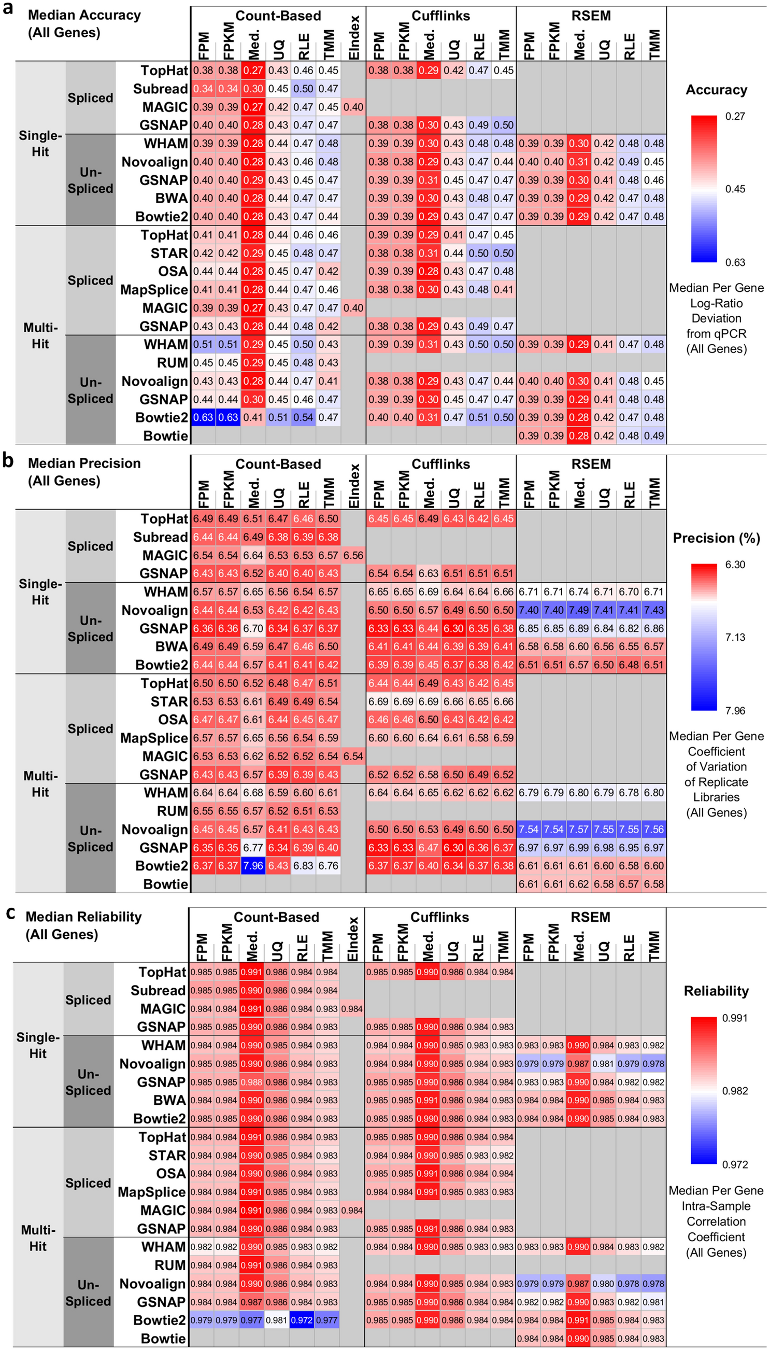
Benchmarking datasets 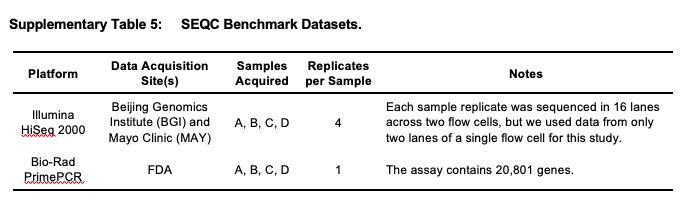
The tools used in pipelines 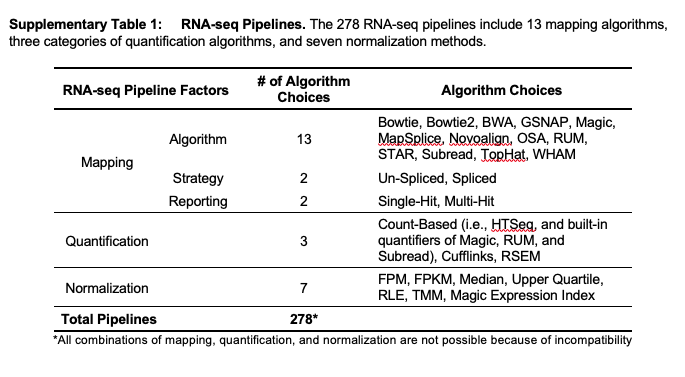
Mapping tools used in pipelines 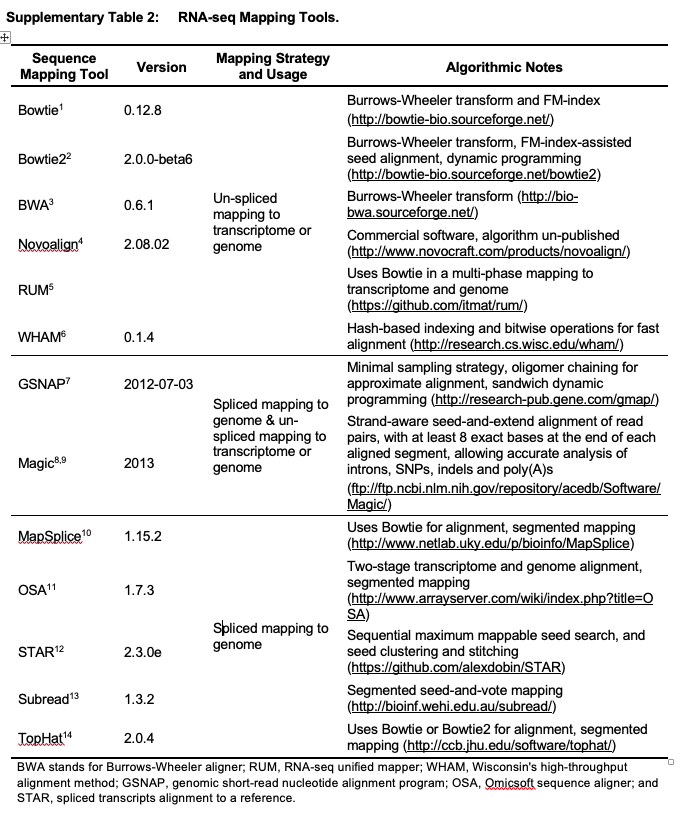
Quantification tools used in pipelines